Heterochromatic domains of DNA account for a large fraction of mammalian genomes and play essential roles in silencing transposons and genes. Heterochromatin loss is associated with cancer, developmental abnormalities, neurodegenerative disease, and premature aging. However, the mechanisms that establish and maintain these domains are not fully understood.
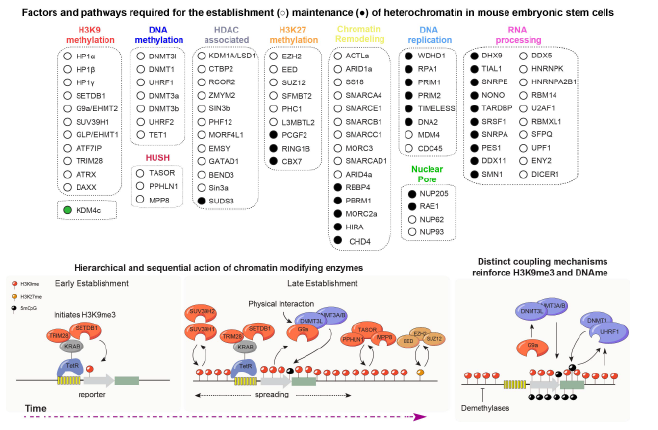
In this study, the Moazed Lab developed an inducible heterochromatin establishment system in mouse embryonic stem cells (mESCs) that allowed the uncoupling of the establishment and maintenance phases of heterochromatin formation. Using this system, they showed that histone H3 lysine 9 tri-methyl (H3K9me3) heterochromatin can be transiently maintained independently of DNA sequence in mESCs and transitions to a remarkably stable state upon differentiation. To systematically identify the pathways required for heterochromatin stability, they performed CRISPR-based genetic screens, which revealed a critical role for DNA methylation in the maintenance of H3K9me3 domains. Genic loci such as imprinted genes behave similarly to the induced heterochromatin locus and are maintained through the coupling of H3K9me3 and DNA methylation, whereas the majority of repetitive and transposon-rich heterochromatin is maintained independently of DNA methylation. The screens also uncovered ordered, non-redundant functions for multiple H3K9 methyltransferases (SETDB1, SUV39H, G9a/GLP), DNA methyltransferases, histone deacetylases, chromatin remodeling complexes, and RNA processing factors.
These findings demonstrate that in the absence of sequence-specific input, newly formed H3K9me3 heterochromatin requires reinforcement by DNA methylation to be heritable. More broadly, the work defines an extensive network of chromatin and RNA pathways that safeguard heterochromatin stability, and provides new insights into mammalian epigenetic inheritance and a valuable resource for future studies of epigenome regulation.