Proteins and metabolites are tightly intertwined in controlling the chemistry of life, yet many of these relationships remain poorly understood. In a new study published in Nature, the Chouchani Lab, together with collaborators at Stanford, Dana-Farber, and Harvard Medical School, introduces a mass spectrometry–based framework that systematically maps functional connections between proteins and metabolites in living tissues.
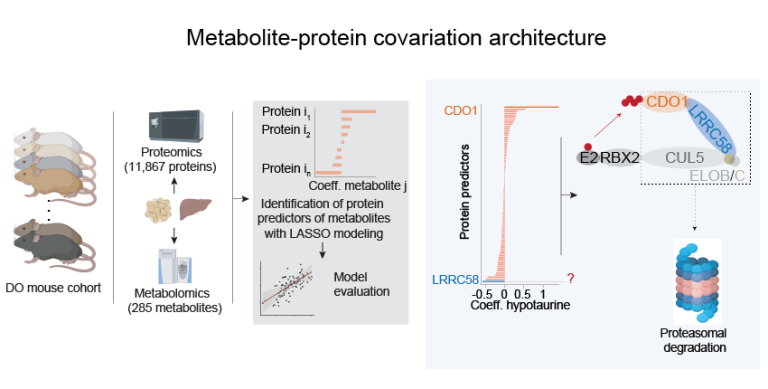
By leveraging genetic diversity in outbred mice, the team developed a resource called Metabolite–Protein Covariation Architecture (MPCA), which nominates thousands of previously unrecognized relationships between proteins and metabolites. Among the discoveries, they highlight a mechanism controlling how cells break down the amino acid cysteine into taurine, a process that directly influences cholesterol balance in the liver.
The researchers found that LRRC58, a previously poorly characterized protein, serves as a substrate adaptor for an E3 ubiquitin ligase complex that regulates the abundance of CDO1, the key enzyme in cysteine catabolism. When cysteine levels are high, CDO1 is stabilized, which promotes the conversion of cysteine into taurine and facilitates cholesterol excretion from the liver. Conversely, when cysteine is scarce, LRRC58 directs the degradation of CDO1, preserving cysteine for other essential functions like glutathione production.
Knockdown of LRRC58 in mouse livers led to higher CDO1 levels, increased taurine flux, and lower hepatic cholesterol. These findings reveal a previously novel cysteine-sensing pathway and suggest new directions for therapeutic strategies aimed at metabolic diseases linked to cholesterol regulation.
MPCA is now publicly available as a community resource (https://mpca-chouchani-lab.dfci.harvard.edu/), offering a platform for uncovering additional protein–metabolite regulatory networks which may open new avenues for understanding how metabolites shape cellular physiology and disease.
Alternative text for main image and teaser image: Simultaneous metabolomics and proteomics in diversity outbred mice was used to generate a list of metabolite-protein correlations. This identified LRRC58 as a part of the CRL5 E3 ubiquitin ligase which targets CDO1 for degradation, thereby regulating cysteine catabolism.