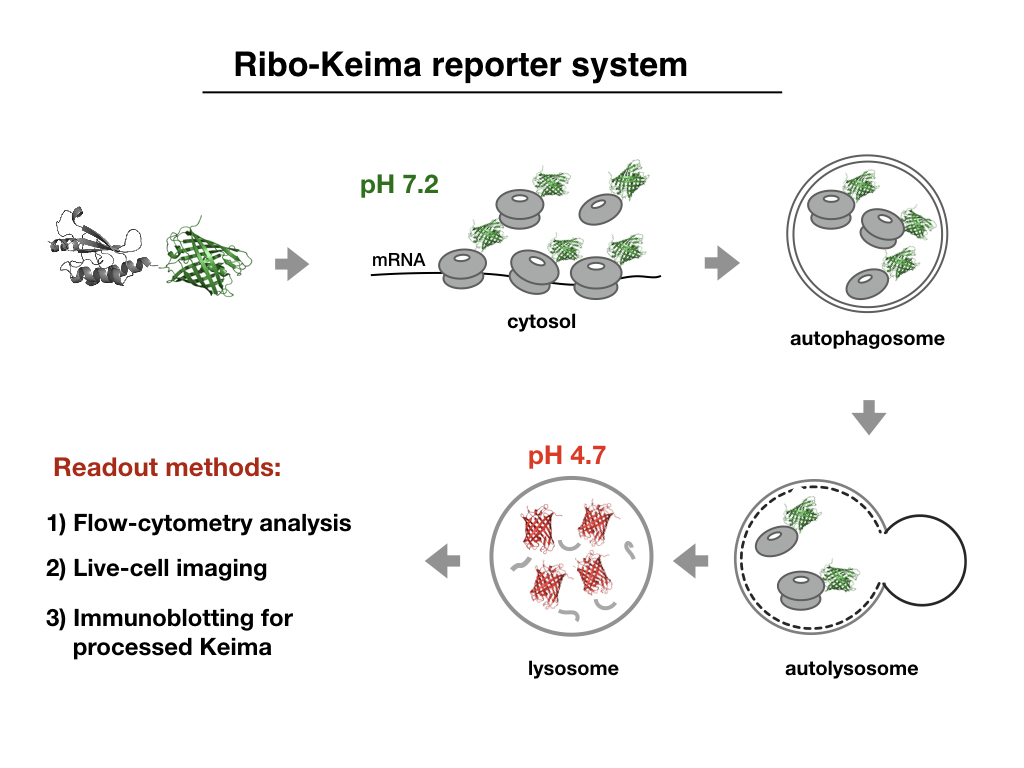
Ribosomes are abundant cellular machines regulated by assembly, supernumerary subunit turnover, and nascent chain quality control mechanisms. Moreover, nitrogen starvation in yeast has been reported to promote selective ribosome delivery to the vacuole in an autophagy conjugation system-dependent manner, a process called "ribophagy". However, whether ribophagy in mammals is selective or regulated has not been examined. Using Ribo-Keima flux reporters, Heeseon An from the Harper lab, in a recent lab report in Nature Cell Biology, found that starvation or mTOR inhibition promotes VPS34-dependent ribophagic flux, which unlike yeast, is largely ATG8 conjugation independent and occurs concomitantly with other cytosolic protein autophagic flux reporters, indicating the absence of selectivity in this process. Ribophagic flux was not induced upon inhibition of translational elongation or nascent chain uncoupling, but was induced in a comparatively selective manner upon proteotoxic stress via arsenite or chromosome mis-segregation dependent upon VPS34 and ATG8 conjugation. Unexpectedly, Heeseon found that agents typically used to induce selective autophagy also promoted increased ribosome and cytosolic protein reporter flux, suggesting significant bulk or "by-stander" autophagy during what is often considered selective autophagy. These results emphasize the importance of monitoring non-specific cargo flux when assessing selective autophagy pathways.