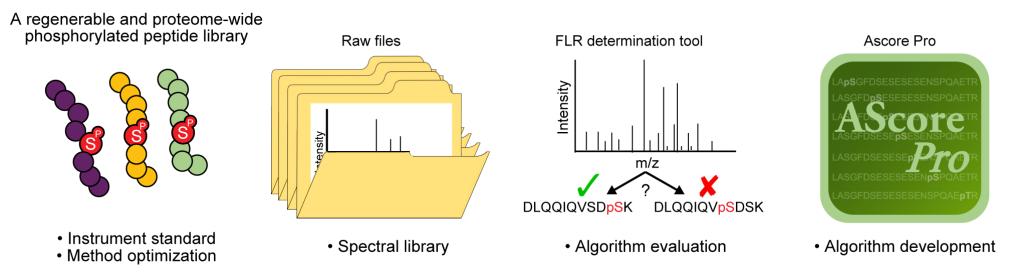
Phosphoproteomics has become an indispensable tool for understanding cell biology. However, to date there have been few tools to standardize and evaluate phosphoproteomics workflows. The Iterative Synthetically Phosphorylated Isomers (iSPI) is a library of >110,000 phosphoserine-containing peptides derived from the observed human phosphoproteome. These phosphopeptides consist of the central phosphoserine residue +/- 15 amino acids from the native protein context and are generated using the Phosphoserine Orthogonal Translation System (pSerOTS). pSerOTS is capable of encoding phosphoserine as a non-standard amino acid at genetically encoded positions in E. coli. iSPI peptides are divided into 10 subpools, with each pool containing unique phosphorylation positional isomers. These features allow for precise knowledge of phosphorylation site position in each pool, as well as low-cost regeneration of the library using standard bacterial culture and protein purification techniques.
One place where the iSPI library is especially useful is in evaluating the methods used to identify phosphopeptides. Though approaches for identifying peptides via tandem mass spectrometry are well established, phosphopeptides have unique properties that make them particularly challenging to identify, and specialized methods are required to ensure that each phosphorylation site is assigned to the correct amino acid within the correct protein. Because the iSPI library contains thousands of phosphopeptides and each position of modification is precisely known, it is ideal for evaluating these specialized methods. Using iSPI to calculate exact error rates, the Gygi lab showed that certain instrument methods caused more frequent mistakes in site identification. Then, using iSPI as a benchmark, they developed an improved algorithm called AScorePro which more accurately identifies phosphorylation sites. Finally, the authors developed a web-based tool (FLR Viewer) that will enable others to use the iSPI library to benchmark their own phosphoproteomics workflows. With the help of the FLR Viewer and AScorePro, the iSPI library will enable rigorous optimization, standardization, and benchmarking of key steps in the phosphoproteomics pipeline and ensure that we map the phosphoproteome with maximal confidence and coverage.
Read the full paper here.