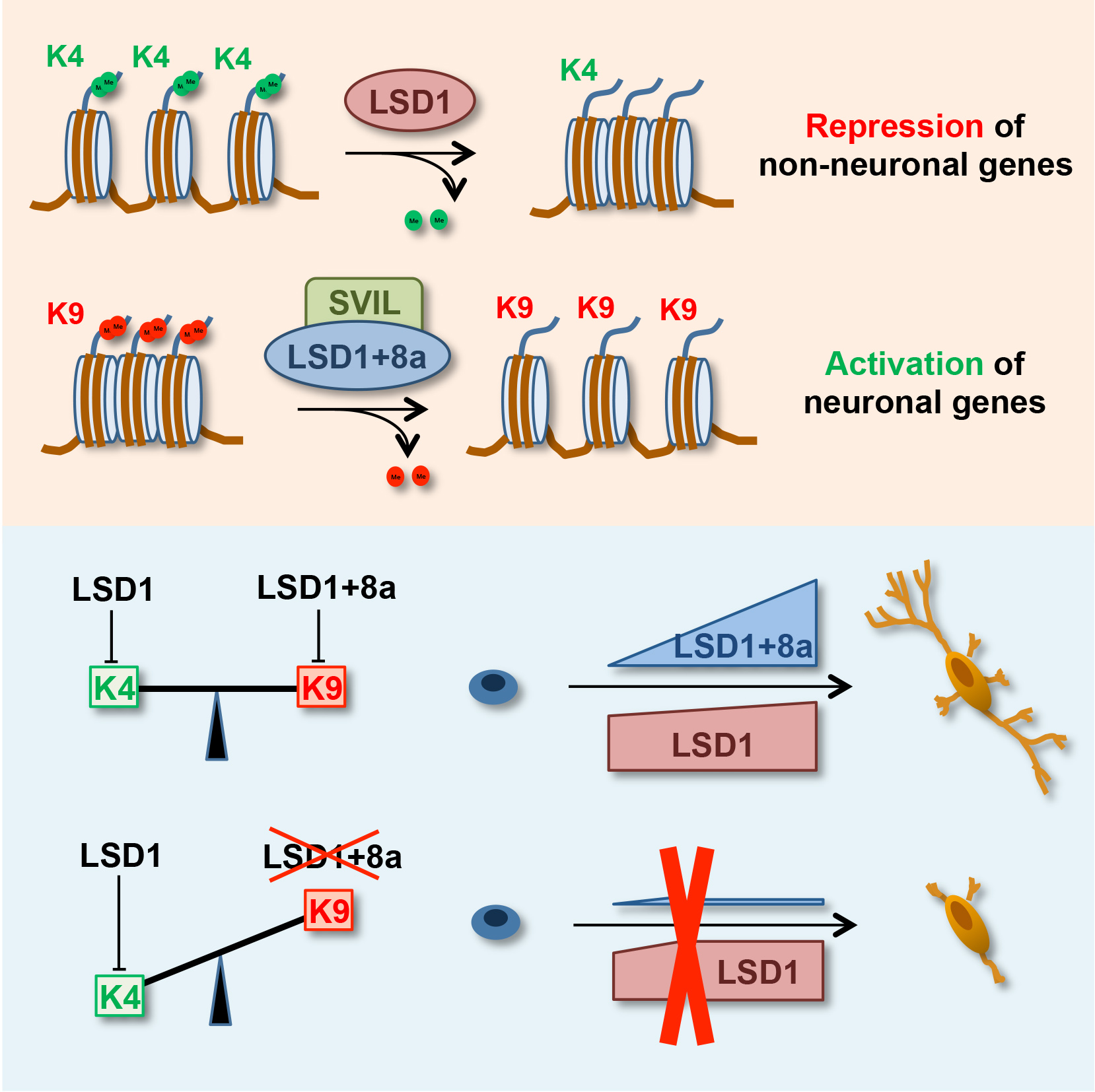
Alternative splicing (AS) contributes to the proteomic diversity. In the brain, it emerges as a pervasive mechanism that plays a crucial role in the regulation of neuron maturation and activity. Chromatin modifying enzymes, that impact chromatin structure and globally control specific gene expression programs, are also subject to AS. However, at molecular level, it is poorly understood how AS can affect enzyme substrate specificity. In a recent publication in Molecular Cell (Laurent B. et al., PMID 25684206), Shi lab shed light on how AS can switch the enzymatic activity of the histone demethylase LSD1. In neurons, AS generates LSD1+8a, a LSD1 isoform containing an additional exon of 4 amino-acids (E8a). LSD1 has been reported to repress gene expression by demethylating histone H3K4. In their study, the authors show that the LSD1+8a isoform does not have the intrinsic capability to demethylate H3K4. Instead, LSD1+8a mediates H3K9 demethylation, in collaboration with the SVIL protein, and activate gene expression at its target promoters. Moreover, LSD1+8a and SVIL knockdowns increase H3K9 methylation levels at their target genes and compromise neuronal differentiation. These findings highlight AS as a means by which LSD1 acquires selective substrate specificities (H3K9 vs H3K4) to differentially control specific gene expression programs in neurons.
Note: This Shi lab publication was commented in its related Molecular Cell issue (Shin J. et al., PMID 25794611).