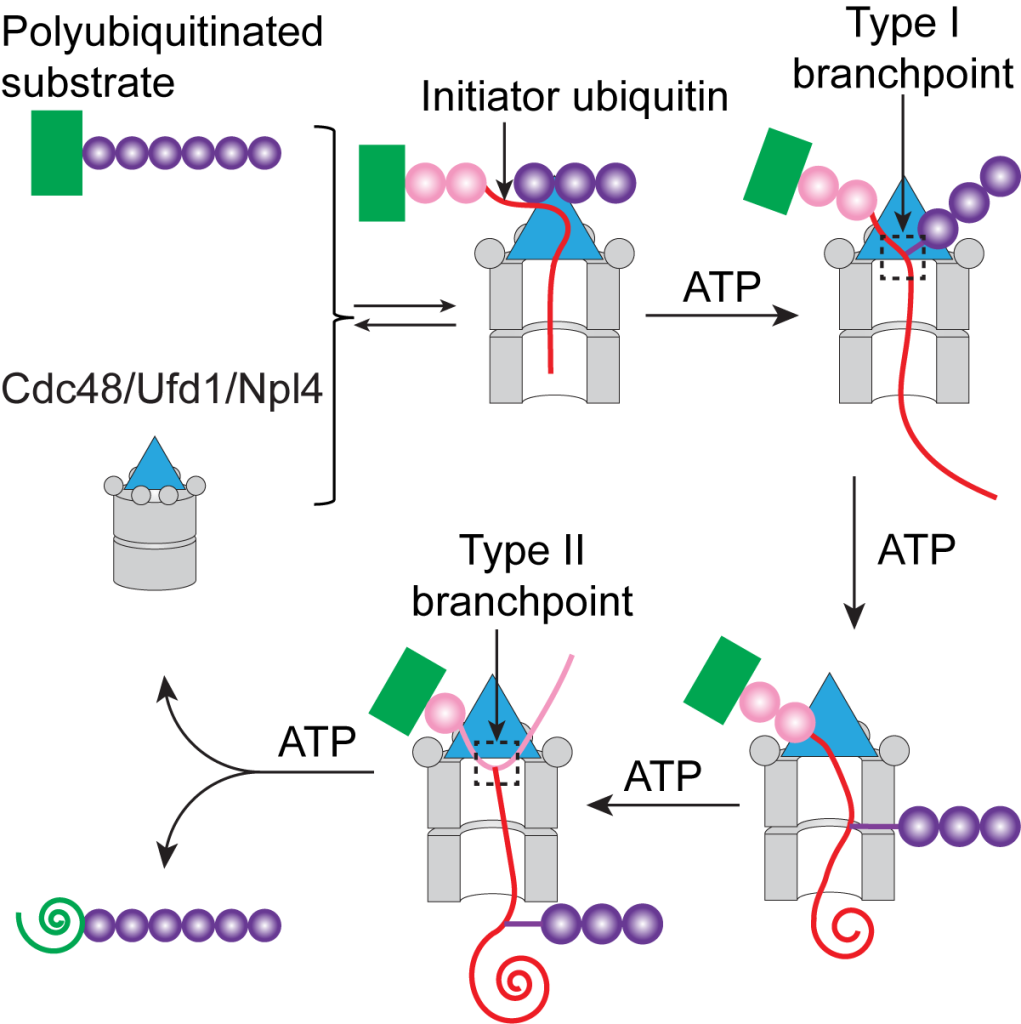
The Cdc48 ATPase (p97 or VCP in mammals) complex extracts polyubiquitinated proteins from membranes or macromolecular complexes for proteasomal degradation. Previously, the Rapoport lab has determined cryo-EM structures of Cdc48 in complex with its Ufd1/Npl4 cofactor and a polyubiquitinated substrate at the initiation state, in which they identified an unfolded ubiquitin. But questions remain as to how the ubiquitin molecule is unfolded and how the Cdc48 ATPase complex unfolds the attached substrate after hydrolyzing ATP. In a recent paper published in Molecular Cell, the Rapoport lab further clarifies the mechanism of substrate processing by showing how the ATPase unfolds the first ubiquitin molecule, how it translocates polypeptides with branchpoints, and how it releases unfolded substrates. They have found that the ubiquitin molecule is unfolded by a cooperative binding to both Cdc48 and the Ufd1/Npl4 cofactor. At the initiation step, the ATPase complex only recognizes the ubiquitin chain but not attached substrate. By hydrolyzing ATP, Cdc48 unfolds polypeptides that are linked C-terminal to the unfolded ubiquitin, including the attached substrate. The distal ubiquitins that are linked to K48 of the unfolded ubiquitin will not be unfolded by the ATPase. At the completion of substrate processing, the unfolded substrate is released from the ATPase without deubiquitination. These findings first explain how a AAA ATPase may translocate through branched polypeptide and greatly deepen our understanding of the role of the Cdc48 ATPase in protein degradation pathway.