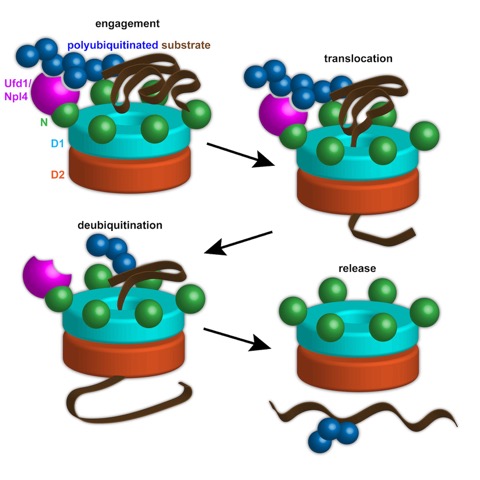
The ubiquitin-proteasome system is responsible for regulated destruction of a wide variety of proteins in eukaryotic cells. Some targets, such as proteins embedded in membranes or stable macromolecular complexes, require prior processing by an ATPase known as Cdc48 in yeast, or p97 in higher organisms. The function of Cdc48/p97 and its cofactors Ufd1 and Npl4 (UN complex) is best understood in the case of endoplasmic reticulum-associated degradation (ERAD), in which Cdc48 extracts proteins from the ER membrane before they are degraded in the cytoplasm. Cdc48/p97 consists of an N-terminal domain that binds UN and two stacked hexameric ATPase rings (D1 and D2) surrounding a central pore. How exactly Cdc48/p97 processes its substrates has been unknown.
In a recent paper in Cell, Bodnar and Rapoport analyze the mechanism of Cdc48/p97. In vitro reconstitution of Cdc48 function with purified yeast components shows that the ATPase cooperates with UN to unfold its polyubiquitinated targets by passing them through its central pore. Translocation requires ATP hydrolysis in the D2 ring, whereas ATP hydrolysis in the D1 ring is required for substrate release from the ATPase complex. Substrate release also requires deubiquitination. Surprisingly, the ubiquitin chain is not completely removed; rather, the remaining oligoubiquitin chain is also translocated through the pore. These results lead to a new paradigm for Cdc48 function in diverse cellular systems.