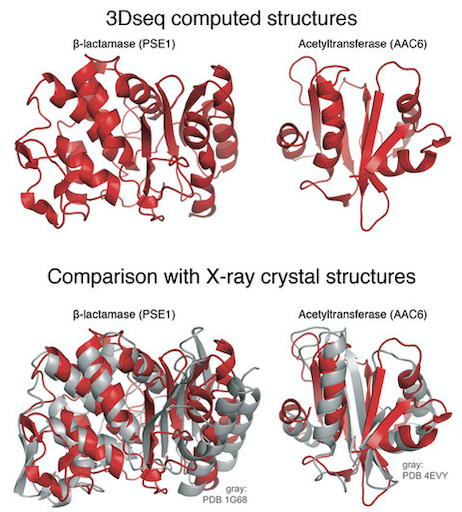
The Sander lab has demonstrated that artificial evolution can provide sufficient information to correctly compute protein 3D structure. Proteins, which are made up of a sequence of amino acids, are fundamental units of biological function and evolution. The key to protein function lies in how these amino acids interact with one another to produce intricate protein 3D structure. In a recent paper published in the journal Cell Systems, the researchers mimicked natural evolution in a laboratory setting by generating hundreds of millions of protein variants and selecting the hundreds of thousands of sequences that retain function. Statistical inference of amino acid interactions from these experimentally evolved sequences enabled them to compute protein structures strikingly similar to those determined by X-ray crystallography. This technology opens the door to a new experimental method for the determination of protein structures - complementary to X-ray crystallography, NMR and cryoEM - and may contribute to a better understanding of the complexities of natural evolution and lead to practical applications in synthetic biology.