Maintaining cellular memory through multiple cell divisions is crucial for development and tissue homeostasis. Part of cellular memory is attributed to the maintenance of silent gene expression states within heterochromatin domains. Previous studies from the Moazed Lab have shown in the fission yeast Schizosaccharomyces pombe, heterochromatin and its-associated histone modifications H3K9me can be epigenetically inherited independently of DNA sequence through multiple cell divisions. Parentally inherited H3K9me during DNA replication is thought to be important for heterochromatin maintenance by serving as the templates for the “read-write” function of histone methyltransferase (Clr4/Suv39h) to restore H3K9me after DNA replication. However, the mechanism by which parental histones are inherited during DNA replication is not understood.
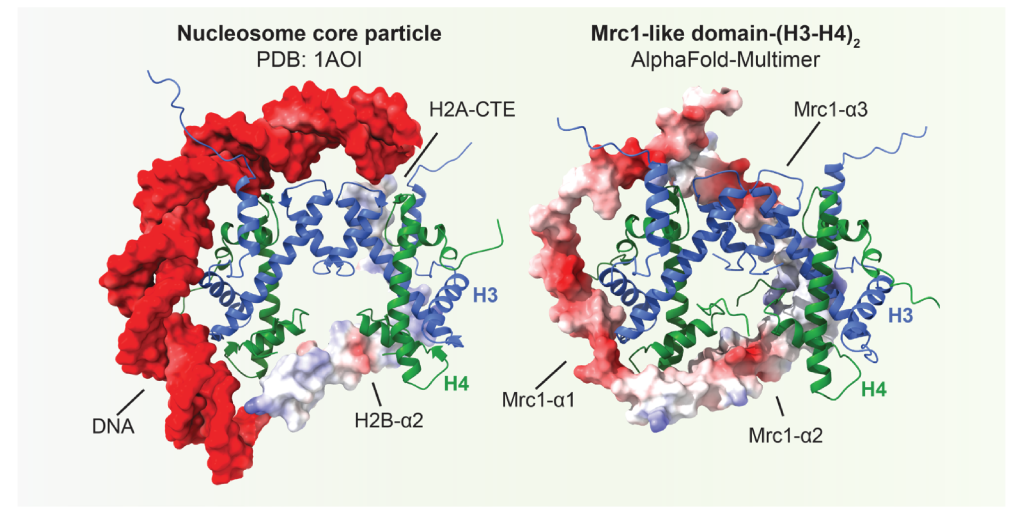
In this study, the Moazed Lab combined AlphaFold-Multimer with in vivo genetics and biochemistry to show that a conserved replisome component Mrc1/CLASPIN contains an H3-H4 tetramer binding domain. The histone binding activity of Mrc1 is required for heterochromatin maintenance independently of its previously known replication checkpoint function. The Mrc1 histone-binding domain structurally resembles components of nucleosomal DNA, H2A and H2B in interacting with the H3-H4 tetramer, suggesting that Mrc1/CLASPIN has evolved nucleosome-like properties to protect the H3-H4 tetramer during DNA replication for inheritance. Through collaborations with Qing Li Lab (Peking University), Songtao Jia Lab (Columbia University) and Zhiguo Zhang Lab (Columbia University), they showed that Mrc1 facilitates the transfer of parental histones to both daughter DNA strands. Both AlphaFold prediction and published cryo-EM structure showed that the Mrc1 histone binding domain interacts with an interface formed by Mcm2/Cdc45 in the replisome, suggesting that the Mrc1 histone binding domain is located at the center of replication fork, ideally positioned to distribute parental histones. Additional AlphaFold predictions also suggest that other replisome histone binding components can dock on various sites within the replisome together with the histone chaperone FACT. This work provides a framework for future studies on the mechanism of DNA replication-coupled parental histone recycling.