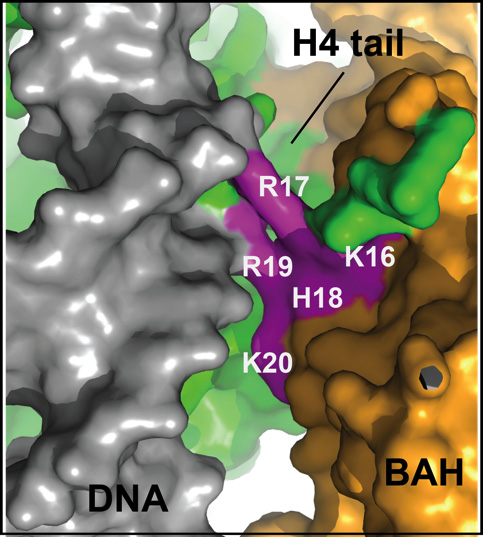
Histones and histone-binding proteins play important role in activating and silencing eukaryotic genes. How binding of factors to the nucleosome may change the properties of chromatin to mediate gene activation or silencing is not understood. A recent crystal structure by Wang et al. (PNAS, 2013) of the nucleosome complexed with the BAH domain of the heterochromain protein Sir3 sheds new light on this question.
Heterochromatic gene silencing in the budding yeast S. cerevisiae requires a nucleosome binding protein, Sir3, and specific histone amino acids, in particular a conserved basic patch region spanning amino acids 16 to 20 in the N-terminal tail of histone H4. Wang and colleagues now provide evidence that association of Sir3 with the nucleosome induces interactions between the N-terminal tail of histone H4 and nucleosomal DNA. The Moazed Lab had previously shown that a conserved region in the N-terminus of Sir3, called the BAH domain, is a nucleosome- and histone tail-binding domain. In this paper, they show that while some histone H4 side chains are critical for binding of Sir3 to the nucleosome, two arginines (R17 and R19), which are critical for silencing in vivo, have no effect on binding of Sir3 to nucleosomes in vitro. To understand the paradoxical requirement for these arginines in gene silencing, the authors determined the 3.1 Å resolution crystal structure of the Sir3-BAH domain in complex with the nucleosome. Consistent with their biochemical data, the structure shows that R17 and R19 point away from the BAH domain and make electrostatic contacts with phosphates of the nucleosomal DNA backbone. In contrast, other histone side chains, which are also critical for silencing, make extensive bonding interactions with the BAH domain. These observations suggest a new role for histone tails in gene silencing and heterochromatin formation, beyond assembly platforms for recruitment of regulatory factors.