Harper Lab charts the structural protein complex landscape of human endosomes
The endosomal system plays a key role in membrane proteome control, which is crucial for proper cell function. Disruption of the endosomal sorting machinery has been associated with neurodegenerative disorders, such as Parkinson’s and Alzheimer’s disease.
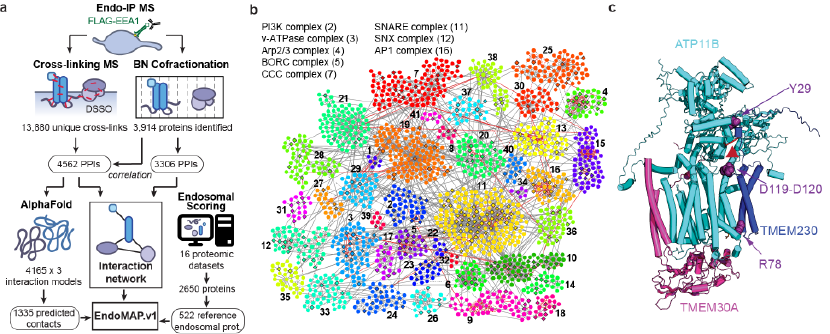
In collaboration with the Walter Lab (HMS BCMP), the Harper Lab has generated EndoMAP, a comprehensive structural protein interactome of human early endosomes.
EndoMAP combines crosslinking and native gel mass spectrometry of purified endosomes with AlphaFold and computational analysis to create a systematic structural interactome of endosomal protein complexes. This resulted in hundreds of structural models for endosomal protein pairs and higher-order complexes supported by experimental data, suggesting structural mechanisms for endosomal regulatory processes. For example, EndoMAP identified new core components of endosomal protein complexes, such as the transmembrane protein TMEM230 as a new subunit of ATP8/ATP11 complex, and TMEM9/TMEM9B as subunits of CLCN3/5 complex. Both complexes were validated by several biochemical and imaging approaches, including observing a loss of interaction in point mutants at the interaction interface between TMEM230 and ATP11B. Given the role of these complexes in neuronal activity, TMEM230 and TMEM9/B were deleted from stem-cell-derived iNeurons to reveal their functional implications on the endosomal proteome.
The complete collection of interactions and 3D models are available online and will serve as a roadmap for merging interactomics with structural biology at scale. For more details, read the full paper here.