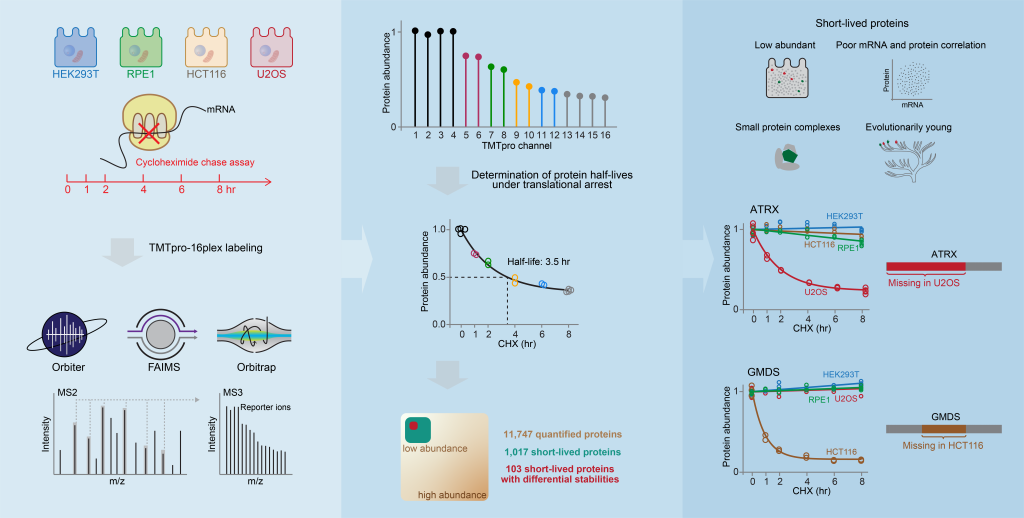
Rapid protein degradation enables cells to quickly modulate protein abundance. Dysregulation of short-lived proteins plays essential roles in disease pathogenesis. A focused map of short-lived proteins remains understudied. Cycloheximide, a translational inhibitor, is widely used in targeted studies to measure degradation kinetics for short-lived proteins. In a recent paper published in Molecular Cell, the Gygi lab combined cycloheximide chase assays with advanced TMTpro-based quantitative proteomics to map short-lived proteins under translational inhibition in four human cell lines. Among 11,747 quantified proteins, 1,017 short-lived proteins (half-lives ≤ 8hr) were identified. These short-lived proteins are less abundant, evolutionarily younger, and less thermally stable than other proteins. The authors further quantified 103 proteins with different stabilities among cell lines and showed that U2OS and HCT116 cells express truncated forms of ATRX and GMDS, respectively, which have lower stability than their full-length counterparts. This study provides a large-scale resource of human short-lived proteins under translational arrest, leading to untapped avenues of protein regulation for therapeutic interventions.