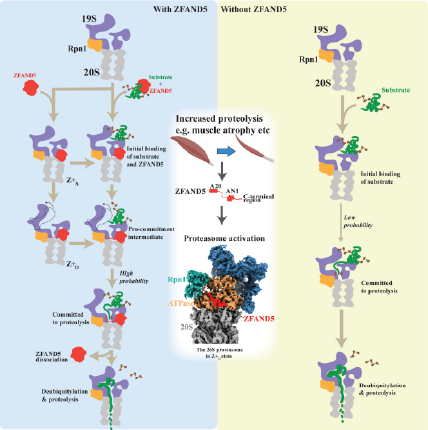
Overall proteolysis increases under highly catabolic conditions or some proteotoxic stimuli, such as starvation, heat shock, or arsenite, in which 26S proteasomal degradation capacity is stimulated. However, the details of the activation mechanism for the 26S proteasome were not fully understood.
In this study, Lee et al., employed multiple approaches, i.e., cryo-EM, cross-linking coupled with mass spectrometry, biochemical assays, and single molecule assays (SMA) and demonstrated new features of activated 26S proteasome. ZFAND5 interacts specifically with Rpt5, Rpt1 and Rpn1 and induces large conformational changes in the regulatory particle (RP) that lead to increased substrate hydrolysis. The degradation assays showed that ZFAND5 substantially stimulates substrate deubiquitylation by Rpn11, which is a commitment step for degradation. Intriguingly, the 19 amino acids sequence from ZFAND5 functions as a peptide activator of 26S proteasome. SMA shows that ZFAND5 binds ubiquitylated substrates, prolongs their association with proteasomes, and increases the likelihood that bound substrates undergo degradation, even though ZFAND5 dissociates before substrate deubiquitylation. These findings can explain the accelerated degradation and suggest how other proteasome activators may stimulate proteolysis.