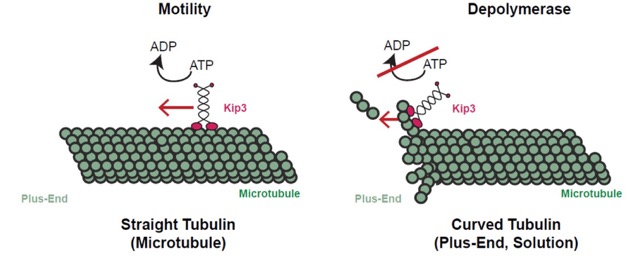
Proper establishment of the size of intracellular microtubule-based structures, the mitotic spindle or the cilia, is key for their cellular function. One class of mechanisms mediating size control of these intracellular structures utilizes molecular motors as “measuring devices”. Kinesin-8 motors have a conserved role in regulating the size of microtubule structures, using length-dependent accumulation at the plus-end to preferentially disassemble long microtubules. Despite extensive study, the kinesin-8 depolymerase mechanism has been debated. In a paper recently published in Developmental Cell, the Pellman lab (with first author Hugo Arellano-Santoyo) defined a tubulin curvature-sensing mechanism for Kip3/kinesin-8 depolymerization. On the straight tubulin of the microtubule lattice, Kip3 behaves like conventional motile kinesin, using ATP for processive stepping, as assayed by single molecule imaging. Upon reaching the curved tubulin of the microtubule plus-end, Kip3 undergoes a switch: Its ATPase activity is suppressed when it binds tightly to the curved conformation of tubulin. This prolongs plus-end binding, stabilizes protofilament curvature, and ultimately promotes microtubule disassembly. This tubulin-binding switch has allowed the co-existence of motility and depolymerase activity in Kip3/kinesin-8s, which is central to their ability to regulate the length of cellular microtubule structures. These findings also illustrate how small scale tuning of binding affinities and rate constants for an enzyme can generate strikingly divergent macroscopic properties.