Obesity is a polygenic disorder with variable penetrance in the general population. Genetic variation is a major factor determining sensitivity to, or protection from, environmental drivers of obesogenesis and metabolic disease. Brown adipose tissue (BAT) is a major regulator of energy expenditure and metabolic physiology due to a specialized proteome that orchestrates futile metabolic cycles, which could be leveraged to treat obesity. However, nearly all mechanistic studies of BAT protein function occur in a single inbred mouse strain, which has limited understanding of generalizable mechanisms of BAT regulation over metabolic physiology.
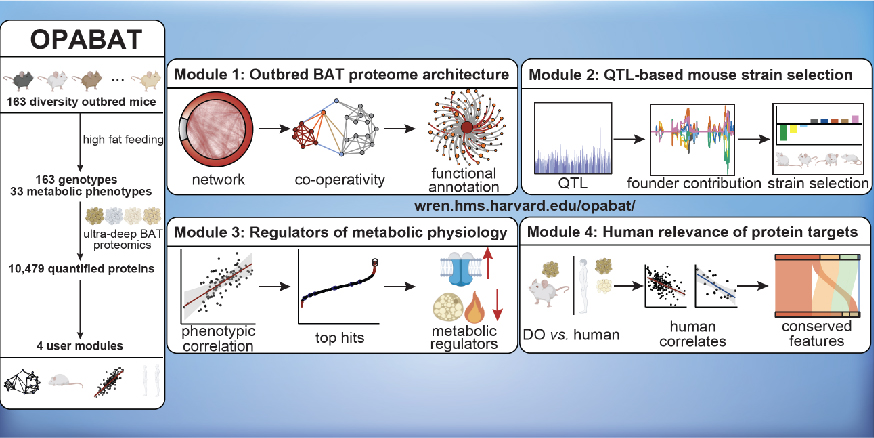
In this study, Xiao et al. performed deep quantitative multiplexed proteomics of BAT across a cohort of 163 genetically defined Diversity Outbred (DO) mice, a model that parallels the genetic and phenotypic variation found in the human population. Leveraging the high variation afforded by this model, they defined the functional architecture of the outbred BAT proteome, comprising 10,479 proteins. Through co-variation network analysis, they assigned novel co-operative functions to 2,578 proteins with 780 established protein networks. This analytic framework enabled systematic discovery of regulators of BAT function, exemplified by the authors uncovering SFXN5 and LETMD1 as modulators of UCP1-dependent thermogenesis. The proteomics measurements were complemented by genomic and phenotypic analyses, allowing the development of a tool to guide selection of mouse strains to model specific phenotypic outcomes. The authors also identified 638 proteins that underlie protection from, or sensitivity to, at least one parameter of metabolic disease. From this basis, they identified the Na+/K+-ATPase α2 subunit as an inhibitor of BAT energy expenditure, which increases adiposity through antagonism of calcium influx-dependent activation of thermogenic effectors. They also collected human phenotypic and molecular data to confirm that many of these newfound protein targets were recapitulate in human populations. This work was a large collaboration across the Chouchani, Gygi, Churchill, and Spiegelman labs, among all twenty labs that participated in this study. This Outbred Proteomic Architecture of BAT (OPABAT – wren.hms.harvard.edu/opabat/) is provided as a resource for understanding conserved mechanisms of BAT regulation over metabolic physiology.
You can read the full paper here.